
- #How to clean sequence in bioedit manual#
- #How to clean sequence in bioedit full#
- #How to clean sequence in bioedit software#
Typically, the obtained raw sequences are provided as input to (multiple) sequence alignment programs. The raw molecular sequence data is produced by DNA sequencing machines which analyze light signals that originate from flurochromes which are attached to nucleotides. Regardless of the concrete (downstream) steps in an analysis pipeline, the precise content and order of nucleotides in a stretch of DNA needs to be determined at the very beginning. In general, molecular sequence analyses involve a multitude of steps which depend on the specific scientific question at hand. Population genetics can be used to infer demographic information such as expansion, migration, mutation and recombination rate in a population, or the location and intensity of selection processes within a genome. Phylogenetic studies can be used to determine how a virus spreads over the globe or to describe major shifts in the diversification rates of plants. Several applications, such as phylogenetic tree reconstruction or inference of population genetic parameters rely on genomic sequence data. Genomic sequence analysis is an important task in bioinformatics and computational biology. Our experiments indicate that estimates of population mutation rates can be affected two- to three-fold by uncorrected errors. Finally, we show that correcting sequencing errors is important, because population genetic and phylogenetic inferences can be misled by MSAs with uncorrected mis-calls. After this pre-filtering, the user only needs to inspect a small number of peaks in every chromatogram to correct sequencing errors.
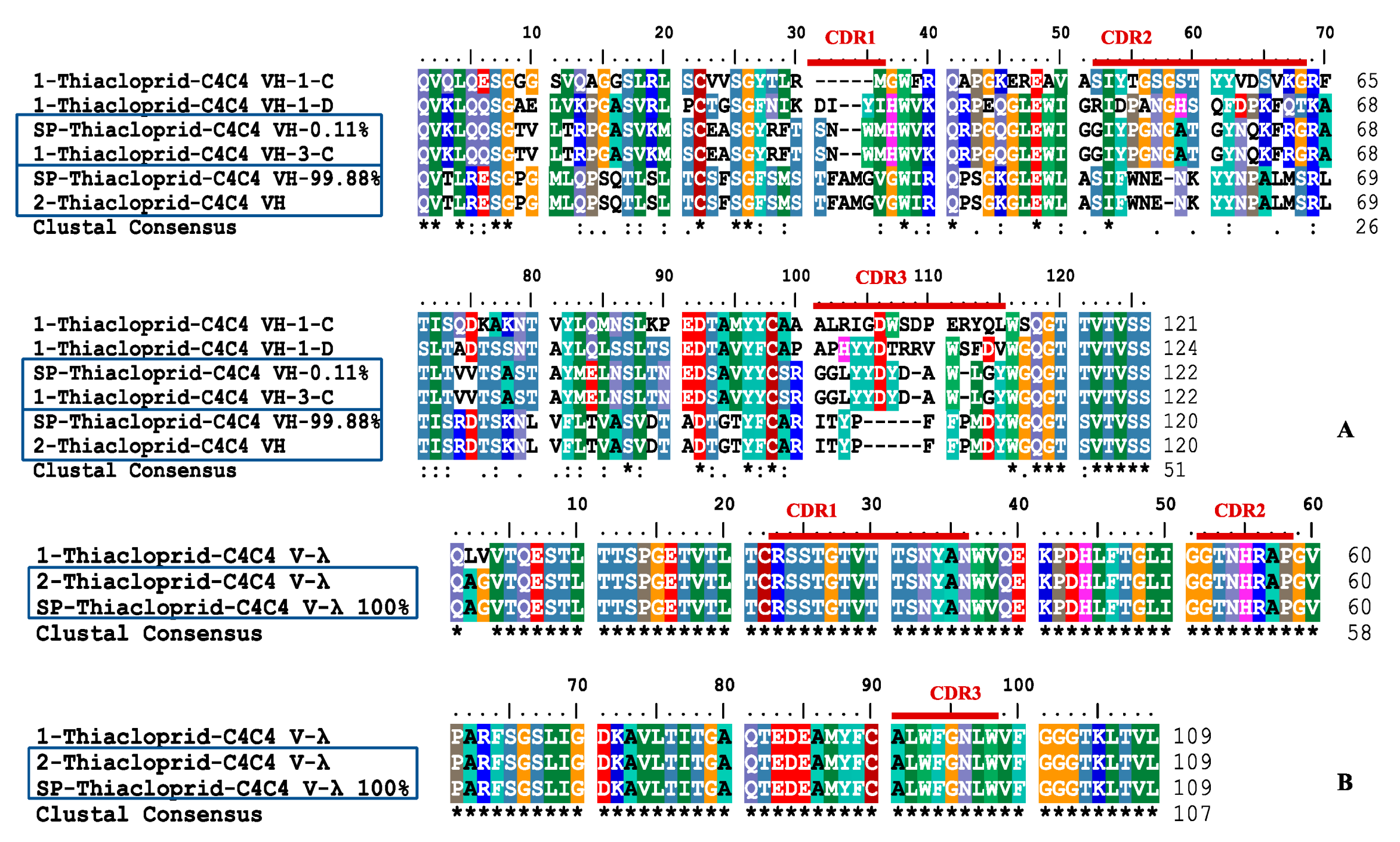
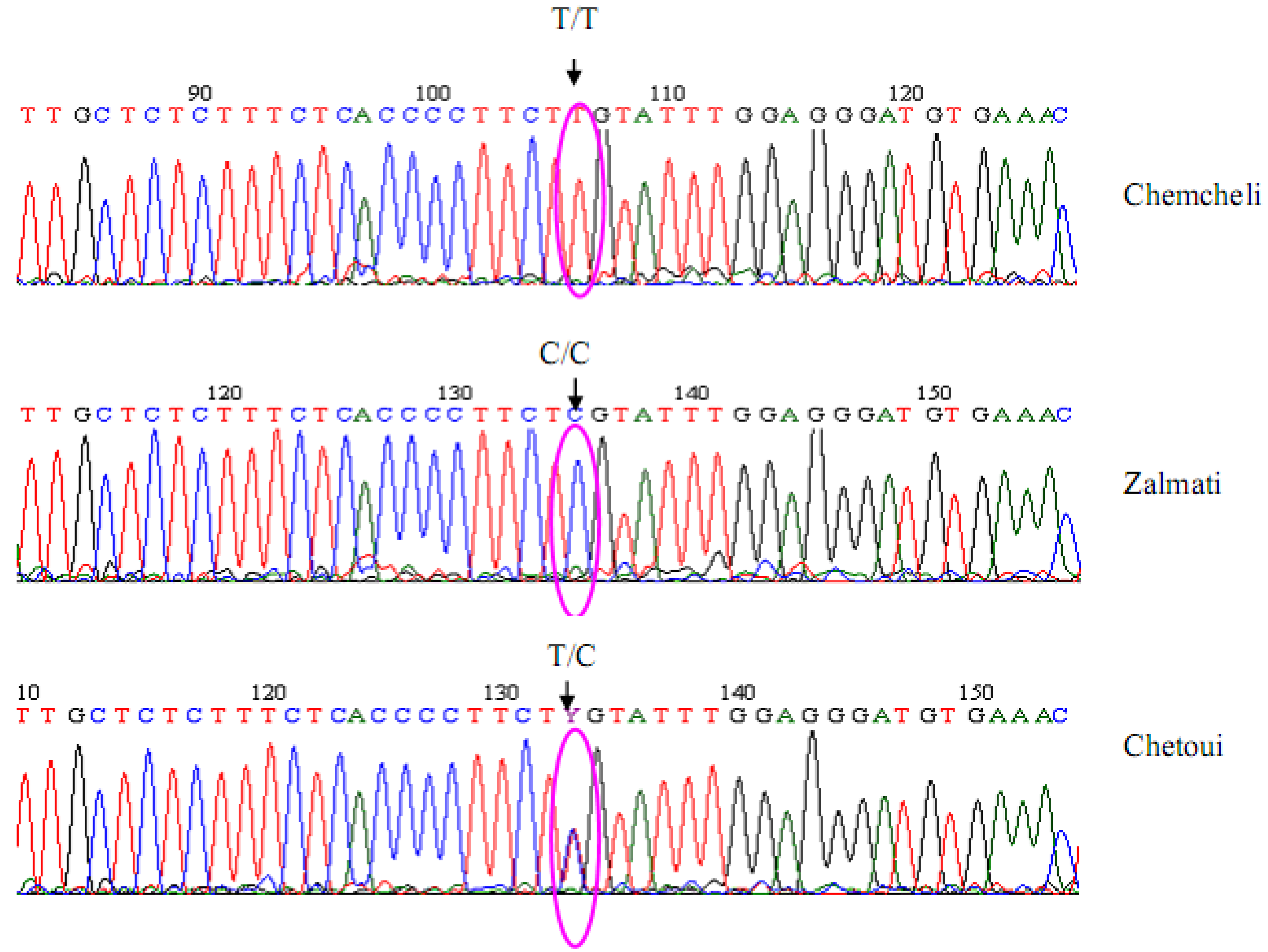
The threshold value represents the sensitivity of the sequencing error detection mechanism. The guided MSA assembly procedure in ChromatoGate detects chromatogram peaks of all characters in an alignment that lead to polymorphic sites, given a user-defined threshold. Initially, the program scans a given multiple sequence alignment (MSA) for potential sequencing errors, assuming that each polymorphic site in the alignment may be attributed to a sequencing error with a certain probability.
#How to clean sequence in bioedit full#
To provide users full control over the error correction process, a fully automated error correction algorithm has not been implemented.
#How to clean sequence in bioedit software#
Here, we introduce ChromatoGate (CG), an open-source software that accelerates and partially automates the inspection of chromatograms and the detection of sequencing errors for bidirectional sequencing runs.

#How to clean sequence in bioedit manual#
As sequence numbers and lengths increase, visual inspection and manual correction of chromatograms and corresponding sequences on a per-peak and per-nucleotide basis becomes an error-prone, time-consuming, and tedious process. Since chromatogram translation programs frequently introduce errors, a manual inspection of the generated sequence data is required. They also generate data that translates the chromatograms into molecular sequences of A, C, G, T, or N (undetermined) characters. Automated DNA sequencers generate chromatograms that contain raw sequencing data.


 0 kommentar(er)
0 kommentar(er)
